Drug Multitask Classification
Using Graph Neural Networks for Drug Classification, Custom built nothing out there like this!
With no end in sight for this Covid-19 issue, there is some time to do crazy ideas. In a previous post I tried to train a neural network that was able to perform algorithms on a given input, which in that cas was cellular automata. There is some refining to be done to get some very good results but overall I saw that training on a certain piece of data, in that case distribution \(P(\{1, 0\}) = \{0.3, 0.7\}\) we were able to scale the algorithm both on the size and variety of other distributions, where we saw that a tiny network with even just\(~1000\) parameters was able to generalise very well on that.
This time around I wanted to play with Graph Neural Networks which are deep algorithms that perform operations on graph data structures. Most of the problems where we use computers in day to day life are structured as graphs and an ability to perform operations on them could be removal of a significant bottle neck. Quick Link: graphdeeplearning.github.io. I am not going to go over what graph, etc. is here I just document my journey to making one.
📦 Finding a Simple Problem
Over time I have realised that 20% part of learning system is just finding the right problem, so I started looking around for a simple dataset and I came across this repo which has some examples and everything and chose to do the Predictive Toxicology Challenge dataset which was small enough and interesting. Here's roughly what the data has to say:
The dataset we selected contains 417 compounds from four types of test animals: MM (male mouse), FM (female mouse), MR (male rat), and FR (female rat). Each compound is with one label selected from {CE, SE, P, E, EE, IS, NE, N}, which stands for Clear Evidence of Carcinogenic Activity (CE), Some Evidence of Carcinogenic Activity (SE), Positive (P), Equivocal (E), Equivocal Evidence of Carcinogenic Activity (EE), Inadequate Study of Carcinogenic Activity (IS), No Evidence of Carcinogenic Activity (NE), and Negative (N).
So I went ahead and downloaded the simple dataset, you get 4 csv files with SMILES (Simplified Molecular Input Line Entry System) notations for the chemicals and it's effect on the particular animal (MM, FM, MR, FR) i.e. whether it was carcinogenic or not. I used pysmiles which takes the smiles string and converts it to networkx graphs. The next step was to convert all this into a single CSV file, which was just 408 lines. Download the CSV File here, there is a minor change that in some of the molecules there is no tests on some animal so the value in that case has been changed to 0.
The next task would be to analyse all the graphs and see the features / attributes and all that. Luckily pysmiles handles all that for us, we can use the build in graph maker and see the molecules.
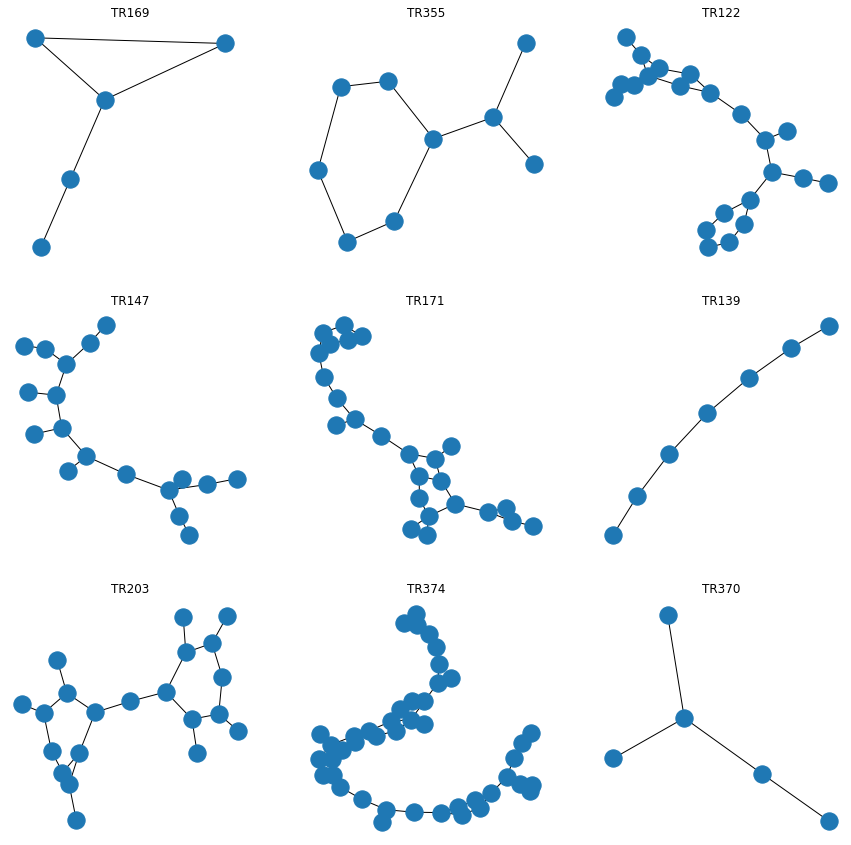
Sample of Molecules
🪐 Finalising the GraphNet structure
There are a tonne of papers and research already done on how to use different types of GraphNets to get good results on this. But we will do our own stuff, taking help from Relational inductive biases, deep learning, and graph networks paper, which establishes the idea of graph network and how all the different pieces are supposed to work.
Each graph has nodes and edges and the information we extract from it can be about the nodes (embedding of the nodes), edges (vector value of the edge connection) or the entire graph (molecule is carginogenic?). The image below will give a better framework for how to think for the graph and the nomenclature around it.

Definition of graph, from here
Thus from the above framework we can say our graphs are \(Undirected,\) \(Attributed\) and we want to extract \(Global\) \(Attribute\) from it. Following are the attributes that we have (📍 means Node and 🔗 means edge):
'Ca', 'In', 'As', 'K', 'Cu', 'Ba', 'Zn', 'C', 'B', 'Te', 'Br', 'F', 'Na', 'Cl', 'N', 'P', 'Sn', 'I', 'S', 'O', 'Pb' convert these to embeddings
1, 0
0, 1, -1 so use this as is
0, 1, 2, 3
1, 1.5, 2, 3
The idea of graph net is to have a general framework for how a single block of network that takes in the graph and returns a graph will look like. It has some of its own operations and user can then chain multiple blocks to get the desired output. Below is the algorithm for it, note that it takes in the full graph tuple \(G(E,V,u)\) and returns \(G(E',V',u')\).

Alogrithm for one block of full graph net, from here
We can now remove and keep pieces that are relevant to us. The beauty here is the composability of the network that can be used in multiple diffrent methods. To start off we do not have a starting global attribute and ony the edges and the vertices. Next up we can return only the new edge and the new vertices, go over this block for a few times and for the last layer return only the global attribute which can then be called the emedding of the graph. This embedding can be used with otehr attributes such as the animal type to get whether some drug is carcinogenic or not.

Composibility of blocks, from here
Okay so we have a rough idea of what we want to see as the network and can now start to build it. Since I have moved over to pytorch from tensorflow, I came across a library called pytorch-geometric, the good thing is that it has a tonne of prebuilt networks ready for us. In full honesty this experiment is just a quick test for an project we are undertaking at my office for using large graph-networks to extract relevant information across large information space. In that we require a full graph-net block in the sense that we need node-level, edge-level and global level information.
🧩 Coding
This section deals with the initial network ideas and coding attempts, saying it was difficult is an understatement. I have added the not-to-do list below, in order to read final code go te below section. But still there are some weird things that I found while working on it. Also note that I am using Encoder-Decoder architecture as that is what looks the most promising right now.
pytorch-geometric has support for the kind of Graph Neural Network Block as described in the above mentioned paper. Though as it turns out the code much more simpler and does not actually need message passing. So I coded up a custom network and acutally did not use the MetaLayer, built the loss functions and bam the training starts. But there is a hiccup, without batching it doesn't actually learn anything.
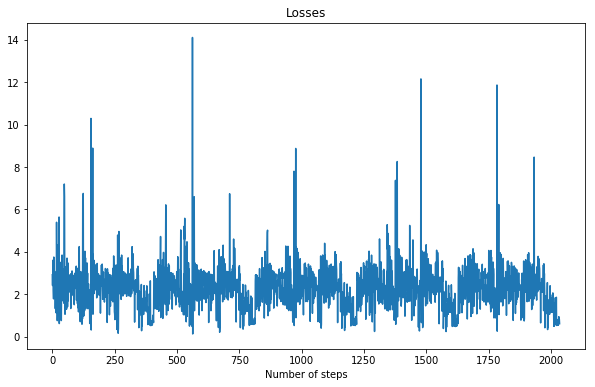
What no batching does, each step is one sample
Batching: This was actually the hard part to do and I did not get some good place to get information on batching. See the challenge with graphs is that the current state of libraries does not allow for such dynamic networks unfortunately otherwise it would hugely progress this field. Adding an image below that tells the difficulty with conventional tensors and I know there is something similar to RaggedTensor in pytorch called NestedTensor but I don't want to get into that.
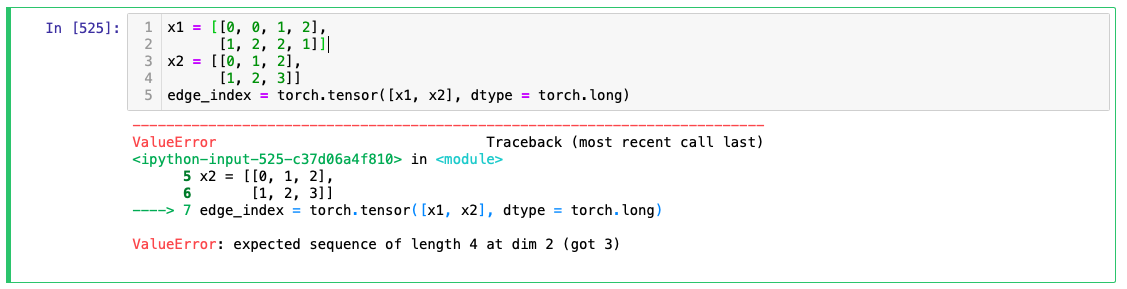
Cannot create tensors with dynamic shapes in pytorch
The conventional method to batch multiple batches to actually combine multiple adjecency matrices as explained here. That should be fine because the we use message passing and there are no overlapping nodes in the two graphs. Say that we have \(n\) batches to sample we get \[\begin{split}\mathbf{A} = \begin{bmatrix} \mathbf{A}_1 & & \\ & \ddots & \\ & & \mathbf{A}_n \end{bmatrix}, \qquad \mathbf{X} = \begin{bmatrix} \mathbf{X}_1 \\ \vdots \\ \mathbf{X}_n \end{bmatrix}, \qquad \mathbf{Y} = \begin{bmatrix} \mathbf{Y}_1 \\ \vdots \\ \mathbf{Y}_n \end{bmatrix}.\end{split}\] Next I go down the stupid way, nice proud of you:
Batching does not guarantee success
Then there are some other things that were a limitation because I was not familiar with pytorch, which include improper gereration of computation graph where I tried to load the embeddings from matrix in the and once an epoch is done set retain_graph = True, which then cause further detoriation of training.
🗿 Coding Success
I haven't added more things here as better explaination is to just read the code, link to gist. Finally I got some success but there were some things I had to giveup, one of them was I had to convert every embedding to one-hot encoding because the batching using DataLoader would not work if there are no tensors and blah blah blah. Long story short I encoded everything to one-hot and the network finally seemed to work like magic.

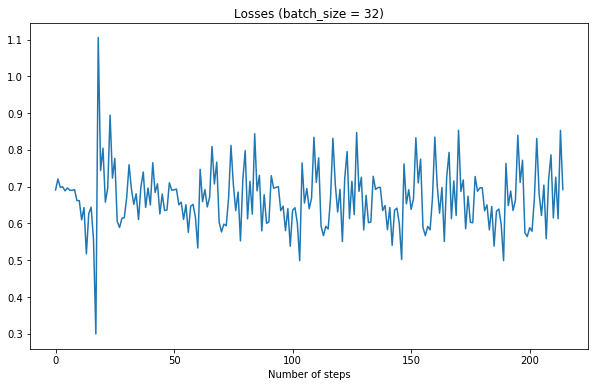
Aah it works, finally. Note the spikes are because graphs are not shuffled every batch
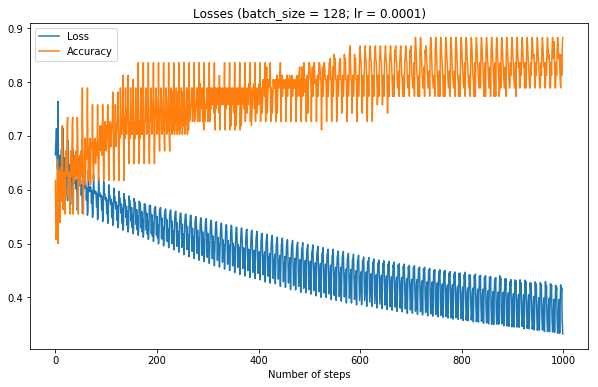
Lowering learning rate creates a nice smooth graph, but note how training takes much longer
Now there are a couple of things I did not implement like testing and benchmarking against the SOTA scores because that was not the point of this at all. I just wanted to make a graph neural network quick.
🚈 Implications
Now that we have one, imagine combining it with previous post where we are able to teach neural network complex rules and algorithms that operate on graphs. That is basically what all these business automation companies do, we can build a comnmon system to just convert all that and build it into an end-to-end AI that operates on algorithms. This can basically allow moving away from large dataset based neural network approaches.
You find something cool or missing do reach out to me at bonde.yash97@gmail.com